User Manual For PlantGSEA
Users for the first time using our website can follow the instructions below.
How to do GSEA analysis |
|
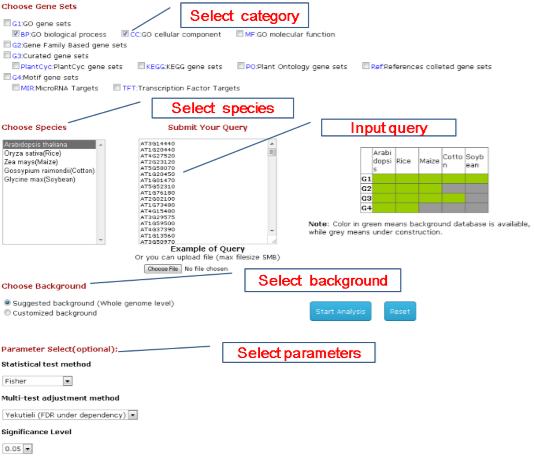
1st step: Choose Parameters of Your Analysis You shoule select categories of gene sets which you are interested in and then the species.In the
query box, you can submit either gene locus ID or affymatrix probeset ID. On the bottom are other parameters
including statistical method and cutoff.
 |
|
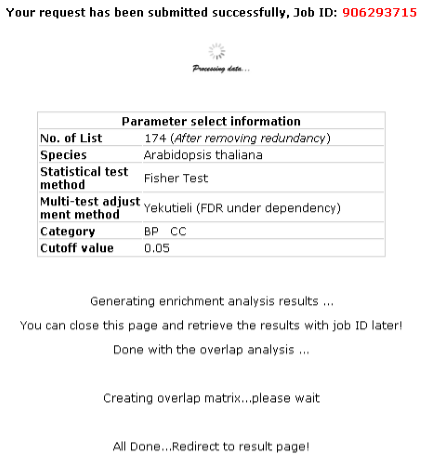
2nd step: Start computing overlap You will get your job ID while the website is running program, which you can retrieve result
even several weeks later.
 |
How to check out analysis results |
GeneSet Enrichment Analysis result is available for download in table-formatted. Page display can be divided into 2 parts.
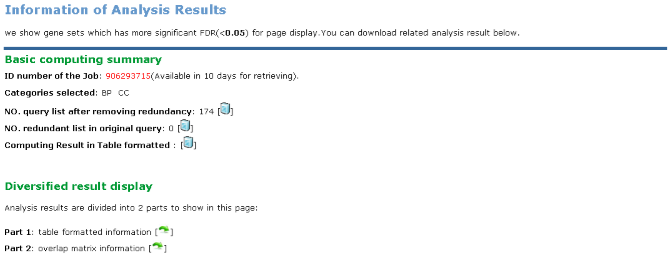 |
|
Part 1: Table Information 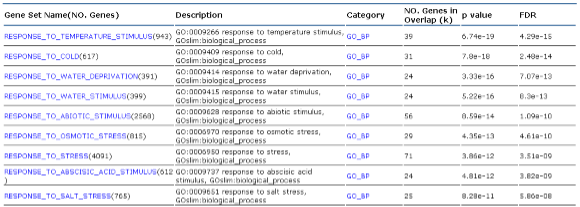 |
|
Part 2: Overlap Matrix Information 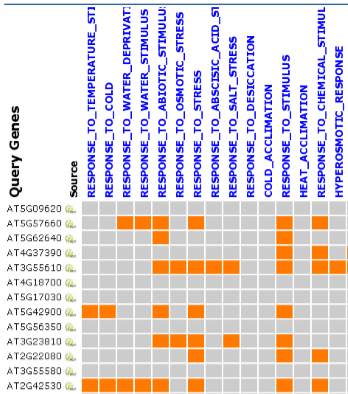 |
|
Part 3: Detail Information 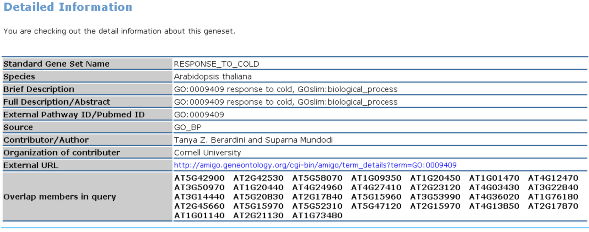 |
|
Part 4: Relationship among significantly enriched gene sets 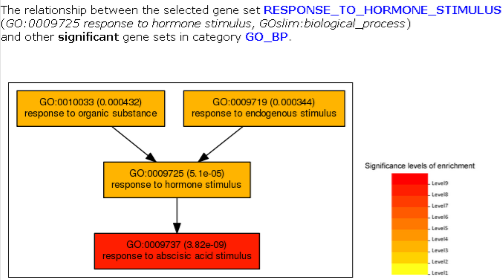 |
How to use Format Conversion Tool |
|
1st : Choose convert type and other parameters You can either submit a list or a text file for conversion between gene locus ID and affymatrix probeset ID.
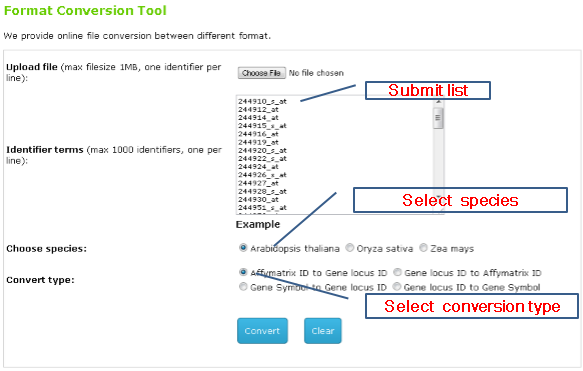 |
|
2nd : You can check out the conversion result on page. 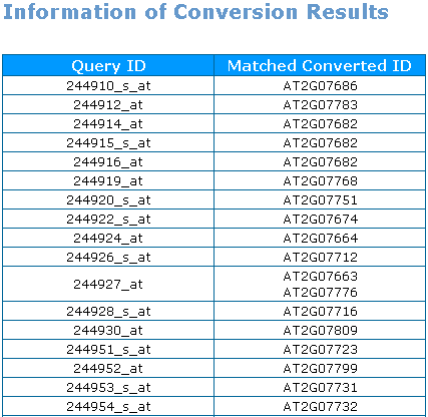 |